cell.codes Lucas Dent | Research Scientist | Computational Cell Biology
I'm Lucas Dent, a Research Scientist at the London Centre for Nanotechnology, University College London. I work in computational cell biology, developing machine learning, informatics and visualisation tools to analyse live and 3D imaging. I also use functional genetics to understand how cells and tissues are built.
Recently, my research has combined imaging with multi-omics (different types of large datasets) and computation to study cell growth, inflammation, geometry, and transcription factor signalling. I also study developmental genetics, focusing on cell proliferation and tissue maintenance. This knowledge is fundamental to human development, regeneration, disease, and creating therapies.
Find out more about some of the biology I work on:
The GTPase regulatory proteins Pix and Git control tissue growth via the Hippo pathway
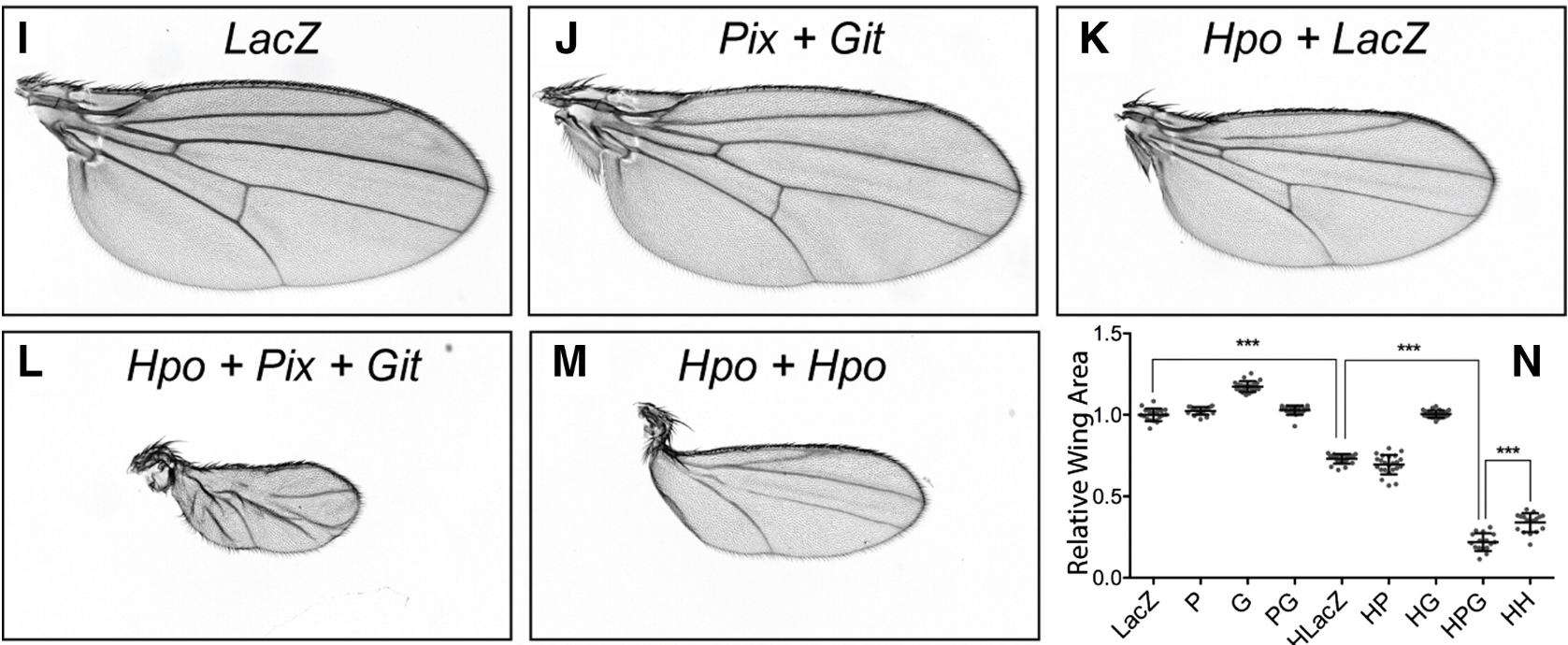
We found two proteins (called ARHGEF7 and GIT1 in humans) that control tissue growth in animals. Here in the picture is an example of how we could change the size of an insect wing.
The dPix-Git complex is essential to coordinate epithelial morphogenesis during development

We found two proteins (called ARHGEF7 and GIT1 in humans) that control tissue integrity and structure. Here is an example of epithelial spheres fusing when we change the level of the proteins.
Informatics approaches to 3D imaging reveal how cells respond to their environment
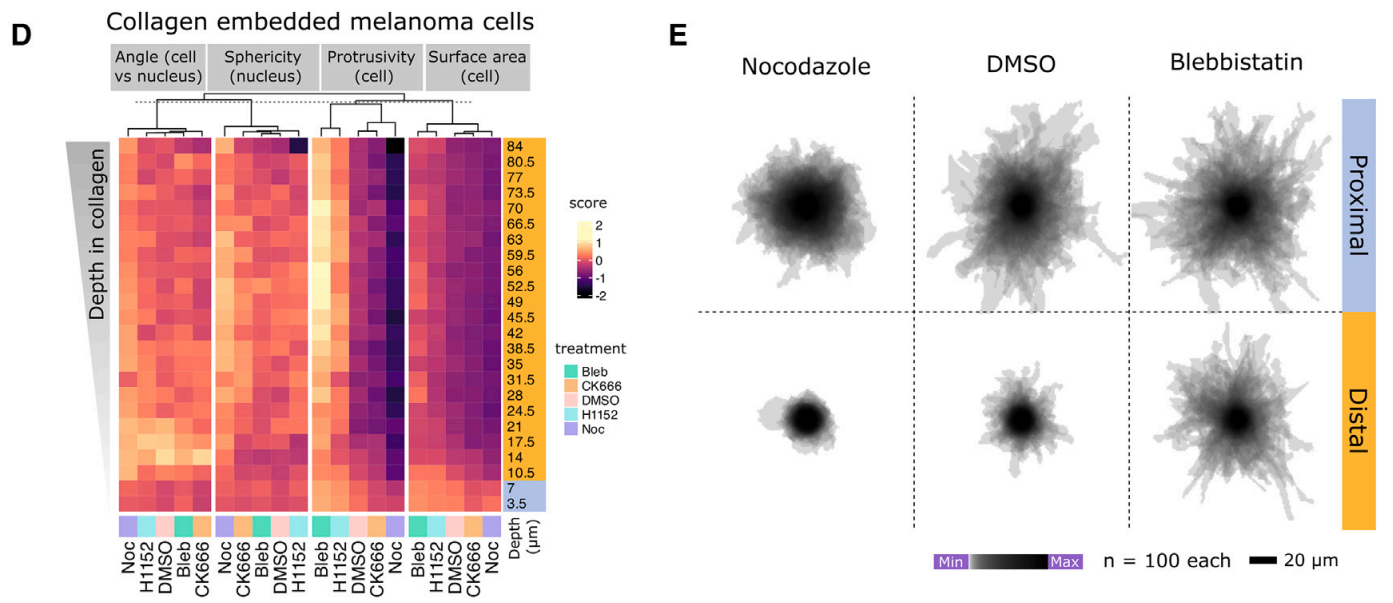
We found cytoskeleton signalling and RhoGTPase controlling proteins (eg. FARP1) control cell shape in response to the geometry and stiffness of the local environment. Here we destroyed microtubules with nocodazole, and you can see they are especially important in soft (distal) environments.
Machine learning approaches to quantify and understand cellular morphology
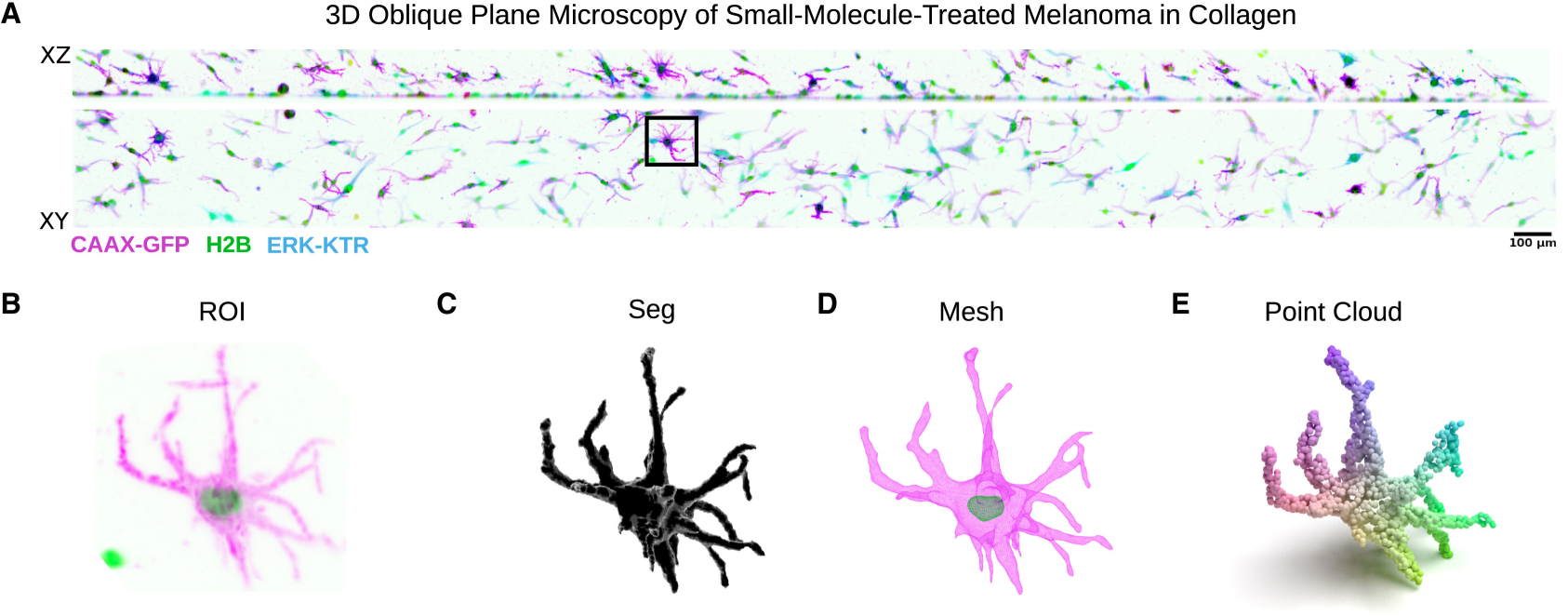
We used geometric deep learning to find out how cell shape signalling changes in thousands of cells, following more than 150 gene knockdowns. Here you can see how we turn images of cells into point clouds, which is the first step in feeding data into our neural network.
Get in touch about science, collaborations, or to say hello. Throw a brick with a note through my window.